Every health institution has in its hands a great amount of patient-level information, such as demographics, medical treatments and conditions, drugs administered and healthcare benefits records. While this increases data available for medical research, one question poses: how can we make use of this information if it derives from multiple sources, where each one has its own structure and language?
A rapid answer would suggest different study definitions for each source, but that would become prohibitively expensive and hinder medical research. Another approach would be to standardize all the data into a common format and only create a study definition to be used across multiple sources. This would still mean an increased overhead in an initial phase to transform the data, but would allow a more scalable research to be conducted.
An example of this common format was specified in the OMOP (Observation Medical Outcomes Partnership) project, specifically in the Common Data Model (CDM) definition, that was embraced by OHDSI (Observational Health Data Sciences and Informatics).
This article will focus on giving a brief overview of OHDSI and its suite of tools and how they can leverage the process of converting the source data to a CDM specification, and, as well, benefit analytically from this.
What is OHDSI?
OHDSI is a health and medical network that focuses its attention on increasing the efficiency and efficacy of research, relying on real observed patient data. The main goal of OHDSI is, therefore, to drive better medical decisions such, for example, in the prescription of drugs, following the idea of a healthcare system that is continuously improving and evolving.
It is based on the principles of open-science, establishing a collaborative open-community across several areas and tools. These tools enable researchers to conduct studies and generate reliable evidence to support subsequent decisions.
Why OMOP CDM?
OMOP CDM specifies a common format from which healthcare data can be converted to, and was based on an initial purpose related to drug safety connections generated from observational data.
The advantages of converting patient-level data to an OMOP CDM specification are clear:
- Only a single definition of a study query is needed, that can be executed across multiple and diverse health providers. This makes research very efficient, transparent, reproducible and scalable.
- Interoperability between health institutions, which allows for easy comparisons.
- Extensive list of standardized Health Concepts and Hierarchies, such as conditions and drugs, as provided by the OHDSI standardized vocabularies.
- Suite of OHDSI tools available in all the processes, comprising tools that range from the beginning of the transformation of source data to large scale analytics and research.
- More health data availability as a community. This is helpful in the case of rare conditions, since more volume of information can be considered. In the beginning of 2022, the OHDSI data network comprised 810 million unique patient records, from 331 data sources.
- The source information is not lost and can be stored in the CDM tables, which allows local and distributed research to be conducted using the same model.
OMOP CDM can be a great way if we are pretending to standardize clinical data, just by looking at the ecosystem of OHDSI tools that can support the whole process.
These tools can broadly be divided into two categories, that depict how source data can be transformed to an OMOP CDM instance and afterwards, how we can extract meaningful information from it: the Extract, Transform and Load (ETL) tools and the Analytics tools.
Tools
ETL
The ETL stage comprises how the tables and fields of a source database are mapped into the Common Data Model. OHDSI provides some tools that can be helpful at accomplishing this work, such as WhiteRabbit, Rabbit In a Hat, Athena and Usagi. The figure below represents the flow of an ETL pipeline representation using the tools referred.
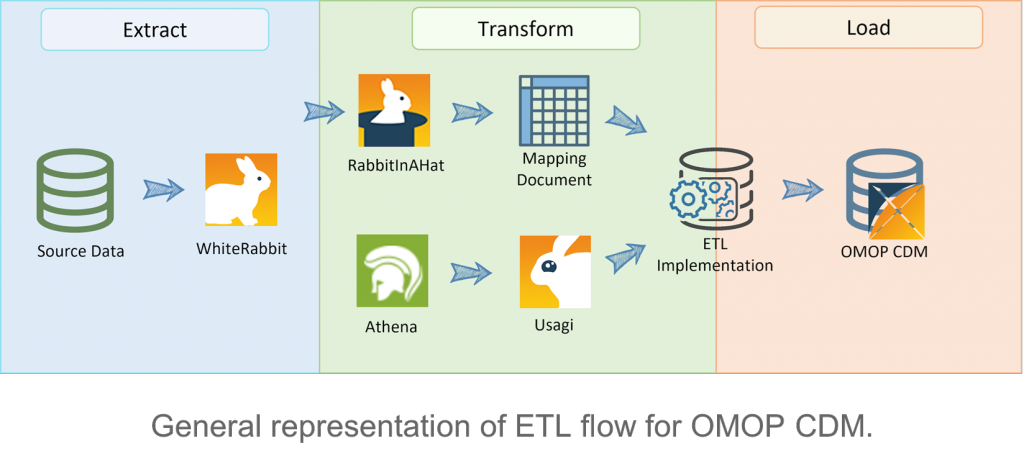
- WhiteRabbit:scans the source database and creates a report with the information associated with each table, while also extracting some simple analytics about the source fields.
- Rabbit In a Hat: relies on the generated report by WhiteRabbit and it is where the mapping between source and target tables is done.
- Athena: used to fetch the Vocabularies. Note that some may require a license, such as the CPT4 vocabulary, for which an UMLS account is needed.
- Usagi: to map source names that do not have a corresponding standard concept, which happens for example when using a specific term in a given language, such as Portuguese. Alongside with a translation mechanism between Portuguese to English, Usagi can be used to find a similar condition in the current concepts and register it in the CDM.
- DataQualityDashboards: Once source data is converted to the OMOP CDM tables, quality metrics can be computed to assure that the process of standardization occurred without errors, for example, to check the plausibility of the values inserted. If the data does not meet the quality criteria expected, some changes need to be made in the previous steps, which places ETL as an iterative process.
Analysis
OHDSI also has open-source tools that are very important in the analysis phase, comprising tools such as Achilles and Atlas.
Achilles provides a detailed characterization of the database and can also be used as a quality check tool. For example, it generates reports with a distribution of the patients or gender, the most occurring conditions, or the data density in the tables, just to name a few from an extensive list of reports that are stored on the same database of the CDM data.
Atlas will be able to generate visualizations based on the Achilles reports, but is also a powerful analysis tool and very useful in research as a method to generate reliable evidence. It allows the definition of cohorts, that specifies how people can be included in a study. With respect to the other functionalities that Atlas offers, to distinguish Population-Level Estimation and Patient-Level Prediction. The first was designed to enable comparisons between cohorts at the treatment level, while the latter tries to predict outcomes from features extracted from the patients, for example, by training Deep Learning models. Moreover, Atlas also provides cohort-based statistics, such as Treatment Pathways, where the sequence of drugs administered for a given type of diagnosis can be represented.
Use cases
OMOP CDM is now more relevant than ever. With the COVID-19 pandemic and the constant mutations of the virus, we soon realized that it was imperative to conduct research in a faster and more scalable way. The Common Data Model paved the way for several COVID-19 studies, which places the standard as a good option for future collaborative analyses.
However, OMOP CDM may not yet be appropriate to all the use cases, since it was especially designed for observational data. Every case is different: some require very specific data to be stored as in the case of medical imaging data, since depending on the case study the number of features of the image and the features itself can vary a lot, which would require changes to be made in the local OMOP CDM tables: instead we could just use the source data, the transformation to a CDM standard would be redundant.
One option could be to add extensions to the OMOP CDM, which is now proposed in the Oncology-CDM extension. The same idea was employed for medical imaging metadata, where DICOM metadata is standardized in a new Common Data Model for Radiology, that can be used alongside the original CDM and the LOINC/RadLex vocabulary to map radiology terminology to the OMOP concepts.
Conclusion
The OHDSI community provides tools for large scale studies to be conducted in the medical world, by making use of real observed patient data, with the goal of guiding better decisions in the future. The OMOP CDM plays an important role in this, since it specifies a standard format for which all types of clinical data can be converted to, leading to a more efficient research.
However, the process of mapping non-standard data to a OMOP CDM format is not linear. This process, known as the ETL stage, varies according to the type of source data that we have and often requires an expert to aid the transformation, which precludes the use of an automatic process that works for all types of data. Nevertheless, OHDSI makes available some open source tools that can make this ETL process less costly.
For analytics, Atlas and Achilles are perhaps the most recognizable OHDSI tools. Achilles provides several insights about the medical data stored, and generates different reports that can be visualized in Atlas. Atlas is not only a visualization tool, but also offers more functionalities, such as different types of Characterization and Population-Level Estimation.
