The digitization of healthcare services is crucial for modernizing and improving patient care at medical institutions. PACS (Picture Archiving and Communication System) archives are the backbone of these services, providing centralized and remote access to images and patient data.
Clinical pathology is just beginning to digitize its services, and many institutions still rely on manual processes. The slow adoption can be attributed to a lack of standardization in pathology imaging and a lack of significant workflow benefits to justify the cost and effort of migration.
It is in this context that the iPATH project proposes a modern digital pathology platform, based entirely on web technologies, named PathoBox. This solution offers tools for the management of digital slides, worklists, medical reports and image analysis in a DICOM compliant network.
Digital image analysis is an emerging field in Digital Pathology with promises of greatly improving the workflow of pathologists. The heterogeneity of digital slides is, however, an obstacle to the development of analysis tools. The different types of tissues combined with different preparation methods creates a wide range of image types that the algorithms cannot cover. This results in very specialized solutions that only work on a very small subset of images.
PathoBox intends to address this issue by proposing a plugin-based architecture for the integration of image analysis algorithms. Image analysis algorithms are installed as modules that grant this architecture the ability to easily integrate new algorithms, without compromising their reusability by other software.
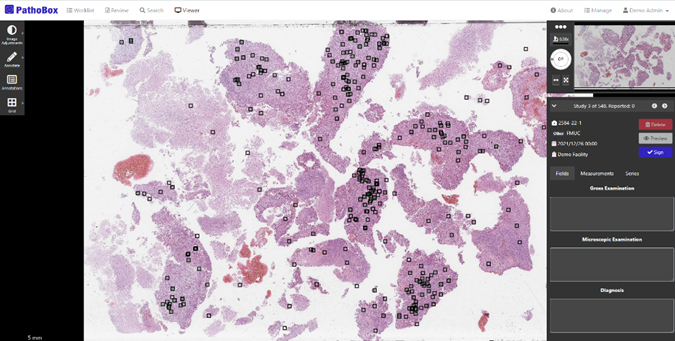
Figure 1: Architecture of the image analysis implementation.
The iPATH project used this architecture to test the integration of a mitosis detection algorithm for the diagnosis of breast cancer. (Figure 2) developed by CCG. The algorithm was trained using annotations validated by the pathologists, produced by the algorithm in an active learning environment. After a few training iterations, the algorithm reached a solid state to be used by the pathologists, and it was integrated into the interface of PathoBox. Pathologists can select a region of the image, using annotations like circles or polygons, to delimit a specific area and send it for analysis. The algorithm will return a list of all the mitosis found in the selection.
Figure 2: Screenshot of the PathoBox application with results of the mitosis detection algorithm.
If you wish to know more about PathoBox and BMD, check out our home page. Stay tuned!
See more about iPATH project.
