Biosignals provide key information for detecting and monitoring individuals’ health problems. However, despite their importance, they still are commonly stored in non-standard formats. For instance, electrocardiogram (ECG) and electroencephalogram (EEG) signals can come from many different sources and exist in various digital formats and sizes, which, in the case of EEGs, can reach many Gigabytes leading to a complex challenge for transmission, management and storage.
At BMD, we have been working to tackle this challenge using a Signal Feeder collector that was designed to allow collecting and storing electronic health signals in a format-agnostic way and in a distributed context (multiple providers and one central remote server).
From a high-level perspective, the Feeder provides of the following features:
- Web application with capability to acquire biosignal files.
- Monitoring of vital and neural signals using the file system.
- Automated format recognition.
- Modular architecture for communication with other services.
- Normalization and anonymization of proprietary signals via a web API.
Also, depending on the type of signal, the Feeder either uploads the normalized files to a central server directly, as is the case of ECGs, that could be hosted in the institution/organization/CardioBox cloud service, or, in the case of EEGs, through other mechanisms such as Dicomization and further DICOM C-STORE to a PACScenter instance.
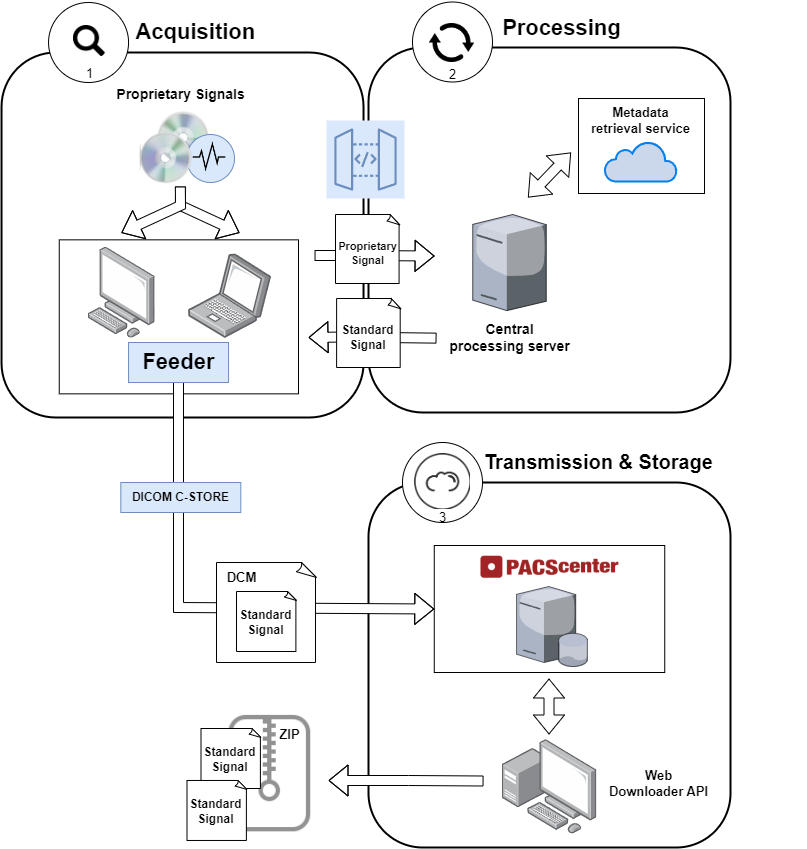
Figure 1 – Health signal architecture workflow for distributing proprietary signals through multiple instances of Feeder to a central PACScenter server
As shown in the Figure 1, using independent central components for processing power that communicate via web API with multiple Feeder instances (that can be geographically distant), it is possible to distribute proprietary signals, which are then normalized and encapsulate into a DICOM file for format-independent storage in a PACScenter server. This combined with a PACScenter extension, the Web Downloader API, it is also possible to fetch and download multiple normalized health signals compressed in a ZIP archive.
This workflow introduces a significant process improvement, as now the raw files containing the signals only have to be moved/copied into a directory monitored by the Feeder, whereby the whole process is automated.
In addition, the Feeder contains a web interface (Feeder UI), accessible by a common browser, which allows users to visually keep tracking of operations, including information about transfers, errors, and the current being processing signals (Figure 2). This is possible since the Feeder stores locally logs of the operations.
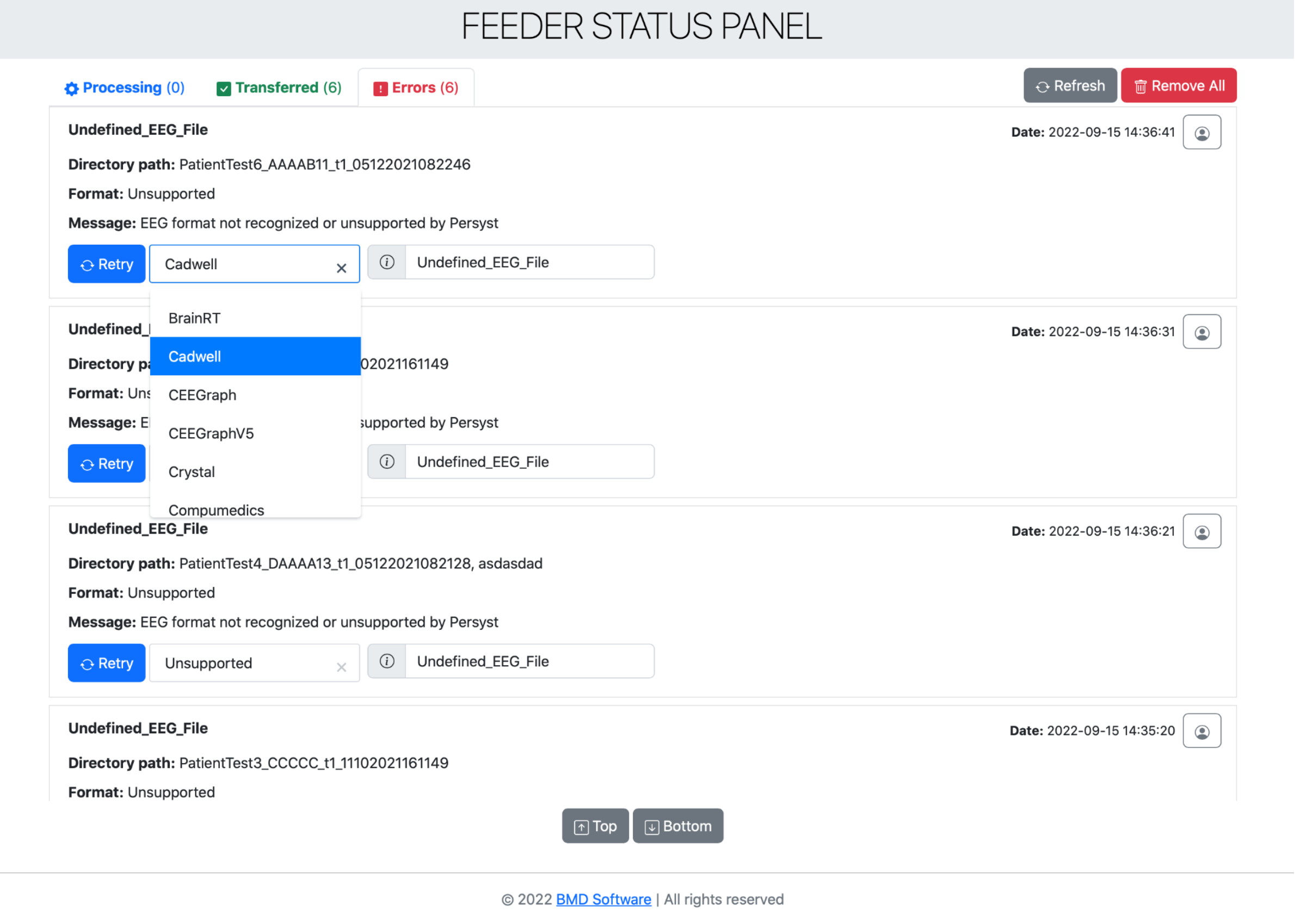
Figure 2 – Feeder UI showing the Errors section.
Moreover, Feeder supports an error detection and handling mechanism which can be used in the UI for retrying the conversion of signals when some error is found during the process (e.g., lost internet connection, processing server down, failure in normalization of a signal).
It is also worth mentioning that the Feeder’s modular architecture allows it to be used by low resource Windows/macOS machines for collecting the signals, since most of the processing power is allocated to a central server.
